The RNA-Targeting Era in Genetic Medicine
By Jim Burns, Ph.D., CEO at Locanabio

In recent years, research focused on correcting disease-causing mutations at the RNA level has made important scientific and clinical advances. RNA-targeted therapies based on antisense oligonucleotides (ASOs) and small interfering RNAs (siRNAs) have emerged with clinical success, proving that messenger RNA (mRNA) can be a valid target for treating genetic diseases. ASOs and siRNAs have produced several drugs, including Vitravene (fomivirsen) for treating cytomegalovirus retinitis, Kynamro® (mipomersen) for the treatment of hypercholesterolemia, Spinraza® (nusinersen) for treatment of spinal muscular atrophy (SMA), and the siRNA drug Onpattro® (patisiran) for treating polyneuropathy associated with transthyretin-mediated amyloidosis. These first generation RNA-targeted therapeutics act by either destroying RNA or modifying its splicing, yet there are significant improvements possible for these approaches and many more genetic diseases that could be treated if there were more effective and adaptable systems to modify disease-causing RNA.
Locanabio is building on the clinical success of ASOs and siRNA by expanding and improving on the current approaches for modifying dysfunctional RNA to treat devastating genetic diseases. We are creating a new class of genetic medicines that can precisely target and modify dysfunctional RNA. Our unique platform uses RNA-binding proteins and engineered effector domains to target and manipulate disease-causing RNA with high specificity across multiple mechanisms of action. Helping pave the way for the in vivo delivery of our RNA-targeting systems are FDA approvals of two in vivo gene therapies using adeno-associated viruses (AAV) – Luxturna® (using AAV2) for treatment of Leber’s congenital amaurosis type 2 and Zolgensma® (using AAV9) for treating SMA in children under two years old. By combining these two validated technologies – RNA modification and gene therapy– we have the potential to treat many more diseases than are possible with ASOs and siRNAs alone. An important advantage of our systems when combined with delivery by gene therapy is that they provide a durable effect after a one-time administration.
The basis of our approach began with research done by Locanabio founder and world leader in RNA biology, Gene Yeo, Ph.D at the University of California, San Diego. Dr. Yeo recognized that gene therapy could be used to deliver RNA directed proteins in vivo which could prevent or treat disease by targeting and modifying dysfunctional RNA. In his initial research, Dr. Yeo and his team showed that the CRISPR-Cas9 system, well established for gene editing, could be mutated so that it binds to RNA instead of DNA. He first applied this new approach, called RNA directed Cas9 (RCas9), as a possible treatment of myotonic dystrophy type 1 (DM1). DM1 is caused by a mutation in the myotonic dystrophy protein kinase (DMPK) gene. This mutation occurs in the non-coding 3’ untranslated region of the gene and leads to a repeat expansion of cytosine-thymine-guanine (CTG) trinucleotide. The expanded CTG is transcribed into CUG (cytosine-uracil-guanine) repeats in the DMPK mRNA which are toxic. Dr. Yeo and his team showed in DM1 patient cells that the RNA CUG repeats, which form toxic foci, could be destroyed by targeting the repeats with his RCas9 system, reversing the biochemical and molecular markers responsible for DM1. More recently, our team at Locanabio in collaboration with Dr. Yeo showed in a DM1 mouse model that the RCas9 system effectively destroyed toxic CUG RNA repeat expansions in muscle and eliminated the molecular markers associated with the CUG RNA toxicity. These data were recently presented at the 25th Annual Meeting of the RNA Society.
In addition to DM1, there are many other diseases caused by nucleotide repeat expansions where use of RNA-binding proteins to direct toxic RNA repeat destruction has potential applications and advantages compared to traditional gene therapy or ASO- and siRNA-based therapies. Some examples include Huntington’s disease and spinocerebellar ataxia type 1 (SCA1), both of which are caused by excessive CAG repeats in the Huntingtin gene and ataxin 1 gene, respectively, and the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), which are caused by the hexanucleotide repeat expansion in the C9orf72 gene.
The RCas9 system has shown great promise in our preclinical research for targeting abnormal RNA repeats and now we have expanded and improved our RNA-binding protein portfolio to include two additional RNA-binding protein systems based on Cas13d and PUF proteins. These new systems provide an advantage over the original RCas9 system as they can be packaged into a single AAV vector. RCas9 proteins are large in size (4.5kB), requiring it to be packaged into two AAV vectors – one for the RCas9 protein and the other for a guide RNA to direct the RCas9 to a target nucleotide sequence. In comparison, the Cas13d and PUF proteins are much smaller (about 3kB and 1.2kB in size, respectively), making it possible to fit either binding protein plus other system components needed to manipulate the target RNA into a single AAV vector. This presents several advantages in terms of dosing, safety, and efficiency. Cas13d is a Class 2 type VI CRISPR system found in prokaryotes as an adaptive immune system against viral infection. Cas13d targets RNA exclusively and can be directed to a specific RNA sequence by a guide RNA. Once the Cas13d binds to the appropriate site, it then cuts the single-stranded RNA. In contrast to the bacterial derived Cas13d binding proteins, PUF (Pumilio and FBF homology) RNA binding proteins are found in most eukaryotes, including humans, and are involved in translational regulation. Our small and human-derived PUFs can be programmed to bind to specific RNA sequences without the need for an additional guide RNA, as is required with Cas proteins.
By coupling these natural Cas13d and PUF RNA binding proteins with various effector domains we can manipulate RNA in ways which provide tremendous optionality to potentially treat more diseases than would be possible by using only one RNA-modifying mechanism (see Figure 1). In addition to being highly effective in targeting and destroying toxic RNA repeats, our systems can be designed to target and destroy non-repeat RNA, presenting a promising approach for treating diseases that do not involve toxic RNA repeats.
FIGURE 1
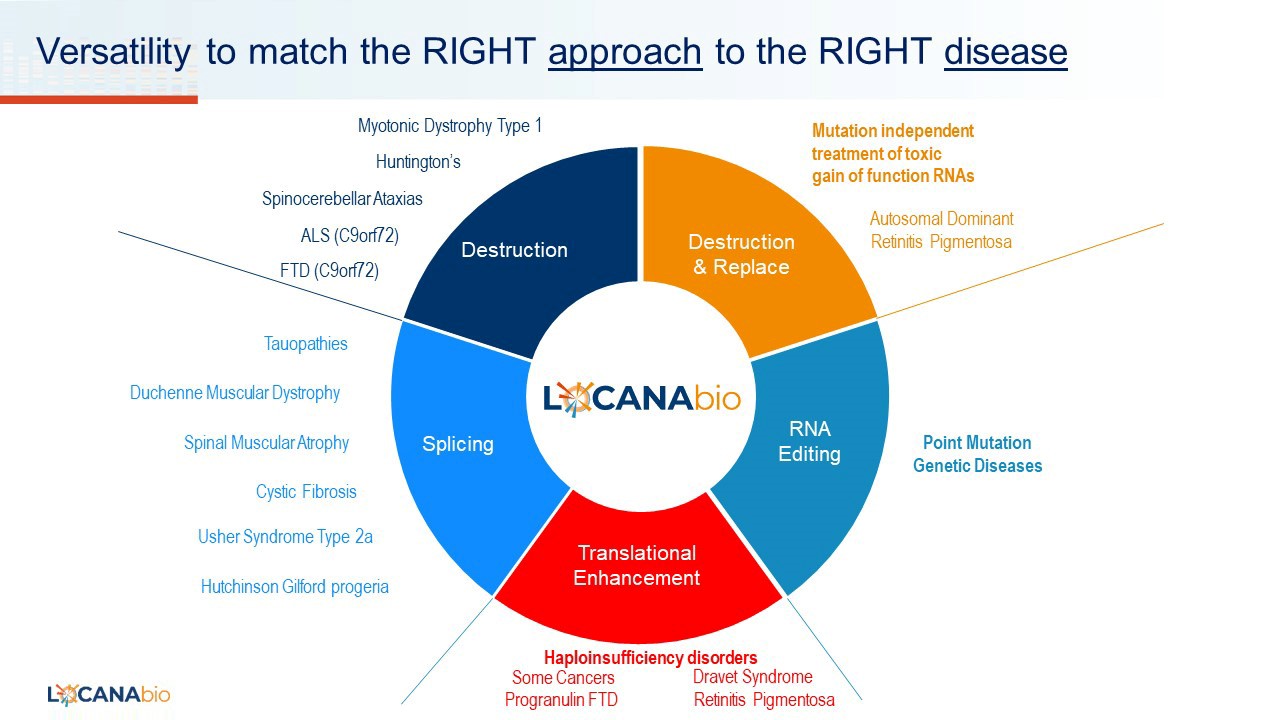
Source: Locanabio
This leads to a very interesting mutation independent “destroy and replace” approach where we can destroy a toxic RNA and within the same AAV vector replace the defective protein with a normal gene. The small size of our PUF systems means that we can easily fit the coding sequences for the binding protein, effector domain, and replacement within the packaging limit of the AAV vector. We are applying the destroy and replace approach to autosomal dominant retinitis pigmentosa, a severe and progressive form of retinal degeneration that leads to blindness. There are more than 150 different mutations in the rhodopsin gene that cause this type of blindness. Our systems have the potential to destroy the mutant rhodopsin mRNA and deliver a normal rhodopsin gene in a single AAV vector that can address all mutations. Finally, our systems are adaptable for also modifying pre-mRNA splicing patterns, enhancing mRNA translation to produce more normal protein levels (as in the case of haploinsufficiency diseases) and RNA editing for correcting point mutations – one of the most common causes of monogenetic diseases.
The breadth of RNA manipulations that our platform provides and the numerous disease types that can potentially be treated is unique in the rapidly evolving field of genetic medicine. We feel that the additional benefit of using gene therapy to deliver a durable effect with a single administration and without changing a cell’s DNA makes our approach very powerful. Building on decades of research, we are hopeful that our novel approach for targeting disease-causing RNA with multiple mechanisms will lead to major advances in treating genetic disease and improving the lives of patients and their families.
About the Author
James (Jim) Burns, Ph.D., joined Locanabio in December 2019 as CEO and Board member. Prior to joining Locanabio, Dr. Burns served as the CEO of Casebia Therapeutics and led the team in discovering and developing new CRISPR/Cas9-based breakthrough therapeutics to treat blood disorders, blindness and heart disease. Prior to Casebia, Dr. Burns spent the bulk of his career at Sanofi-Genzyme, where he held several leadership roles with increasing responsibility, including North America Site Head for R&D where he coordinated R&D operations across key therapeutic areas, and head of Sanofi-Genzyme R&D. Dr. Burns is a former board member of MassBio, a member of the U.S. National Academy of Engineering and a member of the External Advisory Committee for the BioMedical Engineering Department at Boston University. He also served as the industry representative for the Food and Drug Administration General Plastic Surgery Panel. Dr. Burns earned a Bachelor of Science degree in biology from Purdue University and Master of Science and Doctorate degrees in bioengineering from the University of Illinois-Chicago, where his thesis work focused on drug delivery. Following his graduate studies, he was a post-doctoral researcher at the University of Florida.
